





Published on Nov 30, 2023
Image processing and pattern recognition has become a powerful technique in many areas. This includes Engineering, Computer Science, Statistics, Information Science, Physics, Chemistry and Medicine. Anyone who wants to extract data from image or visual project, image processing is required. Most of the image processing software’s are able to process two dimensional images alone. This paper implements three basic image processing operations namely enhancement, blur and segmentation. Each of the operation can be implemented using a variety of algorithms.The algorithms are implemented using and their performance are compared in this paper.The proposed work produces better result of 3D images also.
Keywords: — 2D- two-dimensional ,3D- three-dimensional ,CAD-computer-aided design,CT-computed tomograph , MIP-maximum-intensity projection, MRI-magnetic resonanc image ,ROI-region of interest, STL-Standard Tessellation Language,STLFormat-a file format to the stereolithography computer-aided design (CAD) software created by 3D systems.
Medical images from hospitals consist of a two-dimensional (2D) dataset and provide human body information as a slice, but the human body has three-dimensional (3D) morphology. If we should simulate this 3D morphology, we might be able to obtain more information about the body as well as contribute in the clinical environment to both treatment and surgical outcomes.Although doctors expend a great deal of time and effort in this process, the resultant 3D data is different in each institute. This protocol, therefore, provides standard, easy, and accurate 3D data for clinical fields and even for industrial markets. At present, image data visualization, especially visualization of medical image (such as X-ray, Computer Tomography (CT), Magnetic Resonance Image adjuvant therapy, surgical planning, teaching model, prosthetic design and etc..
However, medical images are usually characterized by faded features utilizing a narrow distribution of gray-levels. Because of this reason, medical image often suffers from high spatial redundancy and low contrast that can be further degraded by the noise introduced in the process of imaging (MRI) and Position Emission Tomography (PET)), has become one of the hotspots of image processing research.As our project required providing the user with a means of segmenting medical images, it was important to find a library that provided a means of doing this, as trying to implement such algorithms ourselves would be difficult, but ITK and VTK provides that. It was essential to be able to display an image, and the results of the segmentation, and so use of a graphics library would be preferable to implementing our own project.
This can be helped to extract quantitative values from images that would be difficult/impossible to analysis directly (eg. brain volume, or even more difficult to measure directly, brain volume change between two scans). This can be used to perform statistical population analyses . Help the radiologist find suspicious things. Radiologists are very good at interpreting images, but they get tired and can miss things. Algorithms can point them to anything that might be interesting (lesions, tumors, etc.)Assist the radiologist in making difficult choices (lung nodule classification, breast lesion classification).
This application can generate complicated mesh 3d model which is very difficult.Medical images from hospitals consist of a 2D dataset, providing information from the human body as sectioned slices. The human body has morphological structure in 3D space. Therefore, to recognize human organs, the 3D reconstruction process is necessary to be reformed using 2D slices. After this, the precise position and shape of organs can be identified. Medical 3D volume imaging is based on unprocessed 3D medical data that contains a variety of medical information. It determines guidelines, standards of medical 3D technology, and the 3D volume image’s safety and quality. This standard describes the generation and practical use of medical 3D modeling for diagnostic and therapeutic applications.
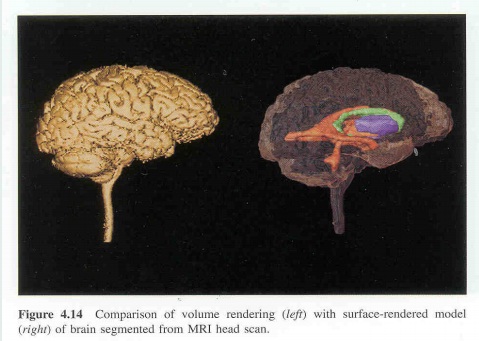
To make 3D models from 2D images, it was necessary to segment the area of interest in the 2D image. 3D models were made by stacking serial 2D images, therefore segmentation was also done consecutively. Generally, segmentation was done on horizontal images, but in some cases, segmentation was done on coronal or sagittal images that were made by stacking horizontal images and then cutting in the coronal or sagittal directions.
3D models shall be made by stacking serial segmented images. Volume 3D models were made as follows: the segmented images were simultaneously expanded onto the next images to build several small volume models, referred to as volume reconstruction. Finally, all volume models were combined. From the combined volume models, a surface model was extracted, which was referred to as the surface reconstruction. Generally, the volume and surface reconstructions were simultaneously performed on reconstruction software. The resulting surface model consisted of the original stacked outlines and numerous triangular surfaces between the outlines.
Filters are intended to process images. They will accept one or more images as input and will produce one or more images as output. We are using mean filter, meadian filter, Recursive Gaussian filter, Binary thresholding filter.
Segmentation of medical images is a challenging task. A myriad of different methods have been proposed and implemented in recent years. In spite of the huge effort invested in this problem, there is no single approach that can generally solve the problem of segmentation for the large variety of image modalities existing today. The most effective segmentation algorithms are obtained by carefully customizing combinations of components. The parameters of these components are tuned for the characteristics of the image modality used as input and the features of the anatomical structure to be segmented.
Surface 3D reconstruction is performed by means of triangulation of a segmented 3D area. The number of triangles determines the quality of the reconstruction. The marching cubes algorithm is the most common method of generating a triangular surface mesh from volumetric data.
STL Format a stereolithography computer-aided design (CAD) software is widely used for rapid prototyping and computer-aided manufacturing. STL files describe only the surface geometry of a 3D object without any representation of color, texture, or other common CAD model attributes. The STL format specifies both ASCII and binary representations. Binary files are more common because they are more compact. The file begins with a solid record (which shall include a name for the object) and ends with an end solid record. Each triangle begins with a facet record and ends with an end facet record.
The current implementation allows more tools to be easily integrated to provide further functionality. Alternative or improved segmentation, skeletonisation, and mesh generation algorithms will be added in the future. Continued experimentation of the current methods may also yield better results.
Overall, more optimisation in both memory usage and computation could greatly improve the program. The large datasets involved in medical imaging push a standard computer to the limits, especially when some algorithms require several copies at the same time. Many of the algorithms do not take advantage of multiple threads which could greatly reduce computation times and allow for more interactive methods. More experimentation with the current algorithms will likely lead to interesting results, which can be used to further develop the tools. Therefore, this project has great potential for future improvements and extensions.
Our project is implemented and demonstrated in visual studio, which uses the c++ code. In this project a novel medical Image processing environment is being developed to import, process and model the 3D images from CT /MRI scanner. A 3D datasets can be loaded into this environment and filtering can be applied to remove the high frequency noise in the image. Next the advance segmentation technique is available for blood vessel segmentation. The developed environment provides the user number of tools for rendering grey scale and segmented datasets. The user can then export the segmented region as a surface model (mesh model) for further processing like printing. Here we take 2D stack of images as a input and we regenerate image as 3D image.
By using 3D data we can easily done the segmentation which very difficult for 2D images .In this work I had to describe and analyze common rendering techniques and their attributes especially under the aspect of how they could be useful in the medical sector.As our project required providing the user with a means of segmenting medical images, it was important to find a library that provided a means of doing this, as trying to implement such algorithms ourselves would be difficult, but ITK and VTK provides that. It was essential to be able to display an image, and the results of the segmentation, and so use of a graphics library would be preferable to implementing our own project.
[1] ABDUL HALIM BIN BABA. image processing learning tool-edge detection bachelor degree. university of technology malaysia 1996.
[2] FIONN MURTAGH. Image Processing data analysis. the multi-scale approach. University of Ulster.
[3] Fundamentals of image processing, hany.farid@dartmouth.edu .(http://www.cs.dartmouth.edu/~farid) [4] JEAN-LUC STARCK, centre d’ etudes de Saclay, Fionn Murtagh. Image processing and data analysis the multiscale approach. University of Ulster.